Here we see the dawn of Homo wentworthi, the only clade of posthuman able to resist the wiles of the AI companions.
Interesting, thanks for sharing! I'm glad my comment was useful to you. I never would've guessed it would cause three genomes to be sequenced...
I'm curious what you think you've learned from this, and how you feel about not being able to experience companionate love? Seems like it might be disappointing in some ways, but also illuminating.
I remember at one point you were looking into trying nasal oxytocin, but it sounds like that wouldn't work anyway. Hopefully in the transhuman future you'll be able to try it out (without totally overwriting your values), if you so wish.
I previously argued for not trying it, but I don't think it would overwrite his values dramatically more or dramatically less than the cluster {weed, candy, sex}.
Which is to say, it might do plenty, but I doubt you need more than a good awareness of your other values so as to get interrupted by them when they'd like you to not have more of [value altering intervention], in order to not get overwritten by introducing a new values glider in your brain.
I would be spooked by a "you feel oxytocin now!" intervention, if one existed and lack of oxytocin really is the root cause, while his rate of potentially future-critical insights is high in ways that seem being-hyperfocused dependent, unless those insights seem to be pointing to "I should apply a values change to get a sense of what those do for {x concrete research relevant reason}", or research output gets stuck on brain patterns that seem to say "I am not being paid in companionate love the way I would like to be, so I'm on strike now" (ie, Burnout<CompanionateLove>).
If I lost my ability to feel companionate love, I would consider that a loss on the order of losing my ability to feel joy. If it's not possible to repair this deficit then so be it, but if it was possible, I would think it urgent to do so. The same way if someone was born without pleasure, you would urge them to fix that, even if they didn't understand why this pleasure thing was so important.
I am definitely skeptical of the idea that we should encourage someone to sacrifice their ability to feel love on the altar of "potentially future-critical insights" they might have. The AI future is too hazy to be demanding such sacrifices at this point. Especially since fixing the oxytocin issue probably wouldn't impact his ability to do good research, unless you'd suggest others should be finding ways to lower their oxytocin to become hyperfocused in the same way.
Never having had the thing is importantly different from losing the thing.
From my perspective, oxytocin makes people do stuff that looks really dumb according to my own values. A central example here is sticking with a romantic partner who sucks up tons of money and/or requires tons of unpleasant emotional maintenance. To my eyes (i.e. without the oxytocin) this seems like a terrible idea. But I see people in those sorts of relationships seeming overall satisfied, presumably because the oxytocin is providing a big positive chunk of value.
That sort of thing does not make me particularly eager to turn on oxytocin signalling for myself, even if I could. It's analogous to heroin, from my perspective: sure, it would change my values so that I feel good while in terrible-according-to-my-current-values situations, but I do not want that.
I think you're underestimating non-emotion-based value of a romantic partner with whom you fuse into an aggregate agent via oxytocin reward, and who is actually able to keep up with your research thread. But not drastically; you recently seem better at filtering for this. I also think you're mostly just seeing what I would call bad relationships and asking why people don't leave them, which may be partly due to oxytocin payout, but I would want to understand your previous relationship better - in ways I doubt your ex would be excited by becoming public, so, not here - before coming to anything like a conclusion that it's oxytocin that is paying for those bad relationships.
On the flipside, I seem to have become averse to romance in a way that makes me think my oxytocin system has been partially suppressed due to repeated updates that it makes me vulnerable to forming bad relationships. It's not clear that that's healthy with respect to my long term value achievement but I don't think it's as obvious as Taylor implies that nobody would ever intentionally give up love for a long period and not regret it. It's still part of me, but is sort of expecting to be payout of success, rather than one of the forms of ape-enjoyableness self-payment that keeps me able to do things. (those are more like "being warm enough" and "enough food". I should probably go pay myself that last one right now!) - I do still get plenty of ambient oxytocin-flavor from companionship with friends who are trying to make a positive impact, though, and I do think I would have Burnout<Oxytocin> if I tried to avoid it entirely.
I think you're conflating your perspective and your values. I think companionate love is not desirable from your perspective, but not necessarily from your values. Think about someone who couldn't feel joy (or pleasure or whatever). They would be saying the same things you're saying now, and they would be wrong, too. Just because a person hasn't felt joy before doesn't mean they don't value joy. I would say if a non-joy-feeling person could take a drug to make them feel joy, they wouldn't be altering their values. Their value function already included a variable for joy; that variable was just always set to zero. Their perspective on joy would change, but they already liked feeling good. Now they just have a new way of feeling good. The desire for "positive experience" is deeper than desire for specific positive feelings you can have, and probably a value you already have.
You could possibly say the same thing about regular people who haven't yet discovered the joy of injecting heroin, but I think the downsides of taking heroin are clear and outweigh the upsides (hence the number of people who regret trying heroin and the dearth of people who think heroin is just awesome). Heroin clearly destroys too much of what you already care about. Maybe you're worried the same thing might be true of companionate love?
Then let me try to sell you on companionate love: It makes me feel like I'm not alone in the world. During times when I've been lonely, I felt purposeless, like any thoughts I had, anything I wrote, anything happening in my life, it all just happened and then faded into the past. But with people I love, I feel like my life has meaning in that it has an impression on them, and their lives have an impression on me, in a way that reduces anxiety and makes life meaningful and full. It makes me feel supported in life. Obviously without companionate love you can still have relationships and practical support, but I'm talking about something more than that. Being a part of something larger than yourself. Caring about more than just yourself (not through morality or empathy or guilt, but actually caring directly). And being cared for in the same way, and feeling that.
The downsides are minimal. Some time, and some obligation to others (which is usually repaid in kind anyway). Most people who can feel companionate love are able to avoid the trap of money-hungry, emotionally-burdensome romantic partners. The people who don't are making an avoidable mistake. (Just make sure the person you love is kind and compassionate before you commit to them. And usually I think it's passionate love/anxious attachment that leads to such mistakes anyway.) I suspect almost nobody on Earth, perhaps not a single person, would give up their ability to feel companionate love just to avoid this trap. Would you give up your ability to feel happiness just so you could free up 5% of your time for working?
Heroin clearly destroys too much of what you already care about. Maybe you're worried the same thing might be true of companionate love?
This is indeed my concern, and I think you are radically underestimating the extent to which oxytocin typically causes people to sacrifice their non-oxytocin values.
For example, about half the population decides to have one or more kids. That's a decision whose emotional motivation is usually mainly oxytocin IIUC. It also, notoriously, typically takes over one's entire life for a decade or more, causing everything else to be thrown under the bus. The whole "midlife crisis" phenomenon is, to my eye, mainly people throwing everything but oxytocin under the bus for a decade, then burning out and needing to relearn to embrace their non-oxytocin values.
Another example: I do not at all buy that most people (or at least most straight males) are able to avoid the trap of money-hungry romantic partners. Here's an infographic:
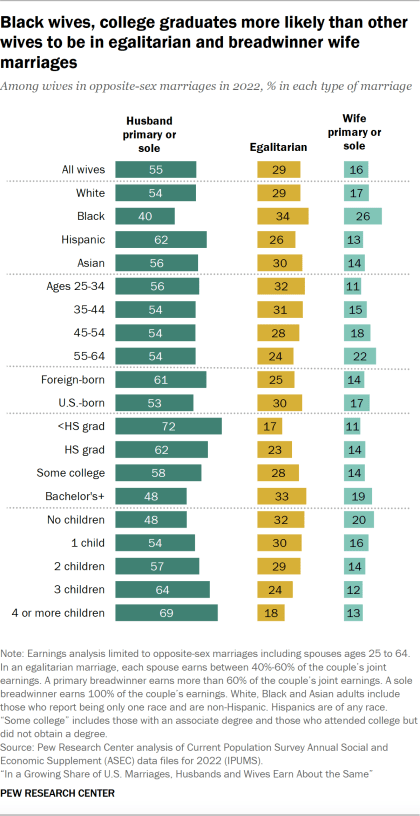
Emotional burden is harder to measure, but it again sure seems to me like a majority of people are pretty darn emotionally burdensome. If someone has even such basic skills as "reliably notice when they're hangry or tired or PMSing and go relax rather than being a pain in the ass", that already puts them in the upper tiers of the population. And without that sort of skill, people are typically deeply unpleasant to be around something like 20% of the time.
A less confident guess, informed more by my own experiences: I suspect that oxytocin typically pushes people to be a lot less ambitious, in general. Most other value-components either satisfice quickly (e.g. food), or push toward a lot of ambition. Oxytocin is one huge value-component which drives people to sink a large fraction of their attention and resources into local things which don't pay off in anything much greater. It's an easier alternative outlet to ambition. People can feel basically-satisfied with their mediocre performance in life so long as they feel that loving connection with the people around them, so they're not very driven to move beyond mediocrity.
That's a decision whose emotional motivation is usually mainly oxytocin IIUC.
I strongly doubt this, especially in men. I suspect it plays a role in promoting attachment to already-born kids but not in deciding to have them.
Oxytocin is one huge value-component which drives people to sink a large fraction of their attention and resources into local things which don't pay off in anything much greater. It's an easier alternative outlet to ambition. People can feel basically-satisfied with their mediocre performance in life so long as they feel that loving connection with the people around them, so they're not very driven to move beyond mediocrity.
I know you are posting on LW which is a skewed audience, but most people are mediocre at most things and are unlikely to achieve great feats according to you, even with more ambition. Having a happy family is quite a reasonable ambition for most people. In fact, it is of the few things an everyday guy can do that “pays off in anything much greater” (i.e. the potential for a long generational line and family legacy).
(Also consider that stereotypically, women are the ones who spend the most effort on domestic and child-related matters, and are also less likely to be on the far right of bell curves.)
I have a nine month old son and in no way feel like everything else is getting thrown under the bus (or that my ambition has decreased as a result).
I do obviously care about keeping my son alive and well, and that does require some degree of compromise with my other goals, but I haven't experienced caring less about my other goals than I did previously.
Also the reasons I care about him have stayed pretty constant both before and after he was actually born, I don't think oxytocin is noticeably influencing how I feel my goals (my guess is it's just causing my feelings of affection towards him in small scale interactions).
Another example: I do not at all buy that most people (or at least most straight males) are able to avoid the trap of money-hungry romantic partners. Here's an infographic:
The infographic you linked does not prove that most people are unable to "avoid the trap of money-hungry romantic partners".
It does not at all preclude romantic partners from deciding together that they want one partner to be a homemaker or take a less highly paid job in order to pursue their interests.
Many people, especially older straight males who make a decent salary, wish to support their partner and have their partner focus primarily on raising their children. They would prefer this to the alternative where their partner works.
The non-egalitarian parts of this graph do not indicate "money-hungriness", nor do the egalitarian parts of the graph clear either part of the couples from being "money-hungry."
It could be true that straight men can't avoid "money-hungry" partners (in fact, most people need money to survive and pursue their goals), but this chart does not at all communicate that.
It does not at all preclude romantic partners from deciding together that they want one partner to be a homemaker or take a less highly paid job in order to pursue their interests.
It can be either, or a part of both.
Many people, especially older straight males who make a decent salary, wish to support their partner and have their partner focus primarily on raising their children. They would prefer this to the alternative where their partner works.
This can be more complicated than it seems. For example, I have much higher salary than my wife. We could easily live all on my salary alone, but we could not keep our current standards on her salary alone.
Therefore, I "choose" to keep my job, because I don't have much of an option. And I would be okay if she stayed at home, because it wouldn't make much of a difference for our family budget, so if it made her happy, why not. Sounds like mostly a free choice I made, doesn't it?
But if you investigate deeper, you could ask, why the difference in our earning abilities? And a part of the answer is that I have spent large parts of my life trying to increase my salary (not perfectly, I made lots of mistakes), while my wife optimized for having a convenient job. And as a result, now I have a well-paying job, and she has an enjoyable job. And by her long-term choices, she helped create this situation where I don't have a choice, but she does.
Even if in our society, women are mostly expected to have a job, the expectations are not the same. Men still grow up expecting the need to have a well-paying job, enough to feed the entire family. For most women, a job is more like a hobby; they expect to pay their own bills and that's it; beyond that, the job is a source of prestige or social contact or meaning. Many men would take meaningless, low-status, socially isolating jobs, if it allowed them to make more money.
So in some sense it is similar to the game of chicken, where the female player already threw her steering wheel out of the window... and the man, seeing that, voluntarily swerves.
Yes, my point was just that the graph alone cannot prove that people aren't able to avoid "money-hungry" spouses and shouldn't be cited as if it does. Arguments comparing it to a game of chicken make more sense to me, especially in child-free marriages, although for most marriages, I think it's important to remember the specific unavoidable burdens on the child-bearing players of the game, a biological "throwing" of the steering wheel out of the window that isn't at all symmetric between players.
I personally am planning to fully support my future spouse and be the primary breadwinner if they're amenable to it, and in doing so have unintentionally opened up a larger amount of potential partners I could end up with. In my particular (majority male) social group, I am far more likely to hear complaints about not being able to find a partner who will be a stay at home parent than I am to find complaints about wanting a partner to pay for half of the bills.
I think it is strictly the better position to be in to be the working one while the other partner is taking care of the home, and am personally too ambitious to be willing to take a low paying job or ever be a stay at home parent, but men who wish to do so are of course fine to live their lives that way too.
I agree that the biological burden is asymmetric. But also, in the past, women used to have about dozen children (most of them died at infancy), while today, it is maybe two on average? From this perspective, women today are more similar to men, than to the women of the past.
I am far more likely to hear complaints about not being able to find a partner who will be a stay at home parent than I am to find complaints about wanting a partner to pay for half of the bills.
I suspect that many of them will find neither. Instead, they will probably find a partner who likes their job too much to stay at home, but not enough to pay for half of the bills (and definitely not enough to let your friends stay at home). Because the job is not optimized to pay the bills.
It's an interesting exercise to take your last paragraph and switch the roles of love and ambition. I wonder if there's anything productive to be explored between those two perspectives, or if those are just terminal value differences.
My issue with unambitious people is that they rarely think more than a few months ahead. This means they end up not developing skills that would make them interesting to talk to and be around, are usually not doing interesting work, and are usually not even doing work well either. Just enough to get by and live a happy life. My other issues with such people is it seems very selfish to choose mediocrity and happiness in the present over happiness for your children, those around you, those you allegedly care about, and even your own future. It also seems rather stupid and inconsistent to say (and believe) things like, "I want enough money to live nicely, without spending 1/3rd of my days working," but then also on every given day take no actions to achieve that goal—just enough to make it through the next few months.
Importantly, for me, you do not have to be ambitious to do these things properly. There are also passionate people who just really enjoy the things they're studying or working on. However, it's harder to get lucky with the right passion, and it's also harder to motivate yourself without passion or ambition, so for most people, a lack of ambition is a serious flaw.
I don't think love (or day-to-day happiness) and ambition are merely different terminal values. I think people who are ambitious in their teens and early twenties can probably experience greater love and happiness for the rest of their lives, so the oxytocin poisoning is actually just making people short-sighted and stupid. It makes sense in an ancestral environment where you could randomly die the next day, but in our modern world we tend to have longer horizons.
I also think John has a visceral reaction of disgust when confronted with unambitious people. I get that. It's the feeling that, "I worked way harder and I didn't complain about the hard work. I still work harder than you, in the hope that I can achieve my goal in a few years. I've even given you advice to help you catch up that I had to struggle to figure out for myself. Give me a break."
I think the downsides are maybe just more visible to you than the upsides. To me, avoiding romantic relationships because I might have to provide 60% of the household's income, or because I have to deal with them being sad or upset sometimes, that seems crazy. (Barring an unreasonable partner, who should be straightforward to avoid. In particular, if you're dating within the LessWrong neurotype, this is less likely to be an issue, and even less so if you're specifically selecting for it.) Likewise, I'd never want to give up love just because I have an hour less a day to spend on other things because I want to spend time with my girlfriend. That hour is valuable to me and worth the break from work.
I think you are right that you'd end up somewhat less ambitious, not that there's a shortage of highly ambitious people who can also feel love. The pattern you pointed to about people being basically satisfied with mediocre performance in life as long as they feel companionate love, I think that's basically a pattern relegated to those who are societal losers anyway, and love is just a consolation prize. It's not like cannabis where it makes you unambitious. I'd guess this trap is most common among the working class, where most adults have no realistic shot of success anyway, so they concern themselves more with relationships and friendships.
I'm kind of saying that you're at least half-right about all your reservations, but that it would be the right decision (IMO) to make the change anyway, if possible. According to your own values, because it's not that you know what you're missing and are choosing not to pursue it according to your values. It's that you don't know what you're missing.
Btw companionate love feels pretty satisfice-able to me. Not sure if anyone else feels differently. But I'm in a steady state with my girlfriend, where I feel the desire to spend a certain percentage of my time with her, and beyond that I feel like I'm satisfied, and she feels similarly. More time together after that is nice in the same way that more snack food might still taste somewhat good even when you're not really hungry anymore.
Think about someone who couldn't feel joy (or pleasure or whatever). They would be saying the same things you're saying now, and they would be wrong
I don't think that this tracks; or, at least, it seems far from being something we can confidently conclude. I don't yet properly understand the type signature of emotions, but they seem to be part of the structure of a mind,[1] rather than outwards-facing interfaces that could be easily factored out (like more straightforward sensory modalities, such as sight). And I think there could be a wide variety of equally valid ways to structure one's mind, including wildly alien ones. I don't think we should go around saying "you must self-modify into this specific type of mind" to people.
Would you give up your ability to feel happiness just so you could free up 5% of your time for working?
FWIW, I'd straight-up turn off all my conscious experience for the next two decades if it made me 5% more productive for those decades and had no other downsides.
- ^
Something like the sensory feedback associated with the dimensions along which a given mind's high-level state/decision-making policy can vary...? Which can be different on a per-mind basis. Doesn't feel like the full story, but maybe something in that direction.
I don't yet properly understand the type signature of emotions, but they seem to be part of the structure of a mind, rather than outwards-facing interfaces that could be easily factored out (like more straightforward sensory modalities, such as sight).
I don't know what you mean by this. You're saying emotions are more fundamental to the mind than sight? I think sight is pretty fundamental. When your mind is randomly stimulated by your brain stem, it's visions (dreams) you experience.
FWIW, I'd straight-up turn off all my conscious experience for the next two decades if it made me 5% more productive for those decades and had no other downsides.
Why?
I absolutely agree that he's missing out on a big thing that, in worlds where he doesn't do, he will eventually regret. The rest of your points are why I mentioned burnout. I would not encourage someone who already feels love to do this, nor would I encourage almost any other researcher to consider their output high enough quality to try to maintain in such a way. Wentworth in particular is one of like 5 people I currently think are consistently making headway on a problem I think we need to solve, and since we seem to have failed to Just Make More Wentworths, I'd rather not lose the one we have due to it turning out he was motivated by an embarrassing thing like "pica for love". Someone who does feel love would probably find themselves resourced by it; it's possible he would too, but https://www.lesswrong.com/posts/Jk9yMXpBLMWNTFLzh/limerence-messes-up-your-rationality-real-bad-yo
So I got my parents’ genomes sequenced. One of them had the frameshift-inducing mutation, as expected. The other had a few substitutions which I share. Alas, that parent's substitutions… were also shared by my other parent
Do you know if anyone ever remarked that your parents are noticeably unusual on the axis of oxytocin-dependent functionality?
"Specifically companionate love - there’s a different hormone (vasopressin) associated e.g. having a crush, new relationship energy, limerence, etc (all of which I do feel)."
This is wrong I think. Where are you getting this info? I can't find any source for vasopressin doing this, and GPT5-Thinking also thinks this is wrong.
I vaguely remember seeing it on wikipedia? Not confident, that whole section was my current gestalt understanding and recollection, and could easily be wrong.
I confess I live on the academic research side, not the consumer medicine side, but the obvious solution to me is "go get a long-read genome so you can phase your mutations." Is that an option? The relevant technology keyword is "PacBio", or given that you already have a short-read assembly, "Nanopore".
Indeed, that will be the next step if and when I decide to invest more effort into nailing this down.
Oxytocin nasal spray is available over-the-counter, including from the official Walmart website. If you want to test this hypothesis, I recommend buying and trying some, and comparing notes with somebody else who's tried it.
Unfortunately, even for people with normal oxytocin function the effects of the nasal spray are typically described as "subtle". I'm moderately skeptical that the nasal delivery route has any effect at all on the usual oxytocin circuits.
That said, I did try some, and had a couple other people try it at the same time. All reports were within-expected-error-bars of "no effect"; nobody experienced anything particularly dramatic.
That's the idea! He could use the nasal spray to figure out for sure how much oxytocin affects him relative to other people. If little to none, that's another line of evidence that the receptors are broken.
Ideally, one would also want to find a volunteer with a typical faculty of love, to try the same spray and compare effects.
If there's anyone who has tried oxytocin, I'd be interested in an account of the experience. The OP moved me to obtain some, but at a dose of 24 IUs I experienced no effect.
If the mutations are shared with both your parents and neither of them share the phenotype, it seems unlikely these are major loss of function mutations, however, it would be interesting to know if anyone else on the side of your family with the frame shift shares this trait with you. If you are at all cost sensitive and have some basic lab proficiency, you could easily spot check the relevant region without long read genome sequencing. Just isolate genomic DNA and PCR up the region. Nanopore sequencing (like plasmidsaurus) can sequence the PCR bands for ~15$ would let you see if the mutations are coming from the frame shift copy or not.
A few months ago, I learned that I probably can’t feel the emotions signalled by oxytocin, the "love hormone". This raises lots of interesting questions - what things I do and don’t feel, how the world looks different from an oxytocin-less perspective, how a lack of oxytocin changes one’s values or goals, etc. But it would be putting the cart before the horse to dive into those questions without first walking through how I learned this about myself and the evidence, so that everybody has an appropriate level of confidence in the underlying assumption.
The Evidence Which Privileged The Hypothesis
It started with investigating a confusion. Lots of the supposedly-happy relationships around me looked pretty awful to my eye, so why the heck were people (apparently) happy with them? What on earth was making these relationships net positive, let alone good?
I wrote a few LessWrong pieces on that confusion, and eventually Caleb Biddulph responded with a hypothesis: perhaps I don’t actually feel much of the thing most commonly called “(companionate) love”, and have therefore been confusing it with something else which I do feel. Caleb also spelled out the physical sensations he experiences with love, and sure enough… his description was not at all familiar to me.
I asked a few other people to describe what companionate love feels like, physically. Sure enough, it did not sound like anything I ever remember feeling.
I had previously asked some people with relationships which seemed bad to me what the major sources of value were from their relationships. Various pointers to “intimacy” topped the list. If that whole intimacy thing was a feeling which I couldn’t experience… as opposed to a cluster of practical benefits, as I’d previously conceptualized it… that sure would explain why these supposedly-happy relationships around me looked pretty awful to my eye!
My ex (of a 10 year relationship) had explicitly hypothesized from time to time that I just didn’t feel love like normal people do. I don’t think either of us had taken that hypothesis completely seriously, but…
Looking back on my childhood, it was clear for a long time that I didn’t form bonds the usual way. I didn’t react the usual way to the deaths of pets. I was eager to get away from my parents at a younger age than normal.
It just made a whole lot of sense.
And physiologically, the obvious guess for what would cause a lack of companionate love was a problem with oxytocin signalling.
Background: Oxytocin
This section is my current gestalt understanding of oxytocin. Take it with a grain of salt.
Oxytocin is often called “the love hormone”. Specifically companionate love - there’s a different hormone (vasopressin) associated e.g. having a crush, new relationship energy, limerence, etc (all of which I do feel). Early work on oxytocin found it released in mothers during breastfeeding, triggering and reinforcing the mother-child emotional bond. Over the years, it’s been associated with lots of other flavors of companionate love.
My current best guess is that oxytocin is the main hormonal signal underlying anything people describe as a feeling of “deep connection”. This includes standard examples of companionate love, like e.g. the love one feels for family. But (I claim) it also includes things like:
The Genetic Evidence
The natural next step was to get my whole genome sequenced, and check my oxytocin and oxytocin receptor genes. I checked the receptor first - it’s a much bigger gene, so a more likely place for a breaking mutation to appear.
Sure enough, there was a single base pair deletion 42 amino acids in to the open reading frame (ORF) of the 389 amino acid protein. That induces a frameshift error, totally messing up the entire rest of the protein. And I did do some basic sanity checks - the sequencing had plenty of depth (i.e. it probably wasn’t noise), and other genes I spot checked did not have any frameshift-inducing mutation.
But that’s not yet a full story. Humans have two copies of each gene, and only one copy had that particular frameshift error. The frameshift error would mess up the protein in a way which triggers nonsense-mediated decay, so the mutated sequence shouldn’t be transcribed much. So by itself, the frameshift mutation should just leave me with 2x less oxytocin receptors than usual (which is usually not a huge deal for signal function), or maybe even closer to normal if there’s any feedback control on receptor density. Upshot of all that: in order for oxytocin signalling to be very broken, there would have to also be some function-breaking mutations on my other copy of the receptor gene.
And there were some other mutations (substitutions, nothing as obvious as a frameshift), a couple of which were predicted by alphafold to be pretty deleterious to the protein’s function…
… but unfortunately today’s standard sequencing technology doesn’t let me know which copy of a gene a mutation is on. We sequence by chopping DNA up into little chunks, sequencing the little chunks, then stitching it all together computationally. But since two copies of the same gene have mostly the same sequence, the stitching step can’t tell which copy a mutation is on, just that it’s on one of them.
The shortest way around this is to get ones’ parents’ genomes also sequenced. If one subset of my mutations appears in one parent, and another subset appears in my other parent… well then, I know that the one subset is on one copy, and the other subset on the other copy (with high probability).
So I got my parents’ genomes sequenced. One of them had the frameshift-inducing mutation, as expected. The other had a few substitutions which I share. Alas, that parent's substitutions… were also shared by my other parent[1]! Which means I can’t fully nail things down with the available information: I don’t know for sure whether the substitutions I have were on the same copy as the frameshift and I have another healthy copy, or if they were on the other copy from the other parent.
That’s my current state of knowledge.
Recap of the key pieces:
Combined with the evidence that made me privilege this whole hypothesis in the first place, I’m pretty confident that my oxytocin signalling is either very weak or entirely absent. But I am relying at least partially on the less-legible evidence which made me privilege the hypothesis; the genetic evidence alone is damn strong evidence in favor of the hypothesis but not fully conclusive on its own.
At this point I wondered if I was using a dubious reference genome, and finding “substitutions” relative to a reference genome which was itself nonstandard. I asked an LLM and its answer was basically “no”, the reference genome is a consensus genome.